Reverse Transcription Polymerase Chain Reaction (RT-PCR)
What is RT-PCR?
The most sensitive technique available for detecting and quantifying messenger RNA (mRNA, or transcript) is RT-PCR, also called Reverse transcription polymerase chain reaction and RNA PCR. The technique is so sensitive that evaluation of transcript from a single cell is possible.
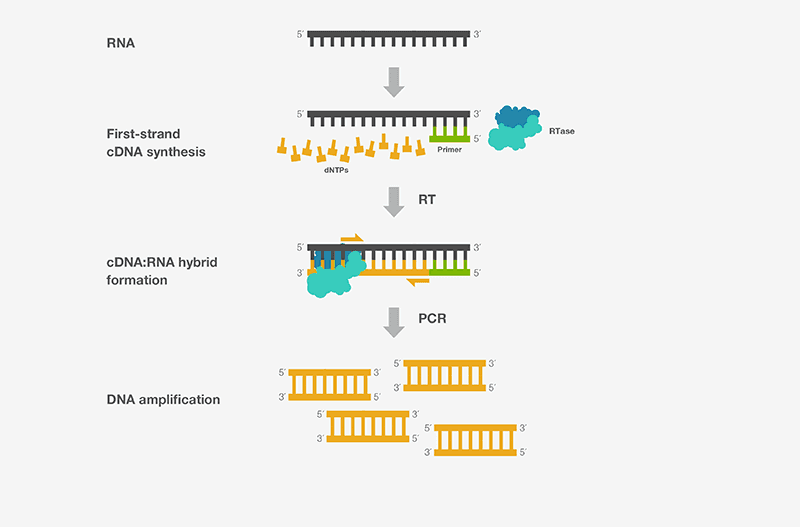
The method uses an enzyme called reverse transcriptase (proper name is RNA dependent DNA polymerase) to synthesize a complementary strand of DNA (complementary DNA, or cDNA) from an RNA template. The resulting cDNA is then used as template in a PCR assay using DNA polymerase. Most clinical microbiology laboratories use RT-PCR assays to detect RNA viruses from clinical specimens.
High-quality template RNA is important for RT-PCR. Numerous kits are available on the commercial market for extraction of RNA from clinical specimens, including specific kits for viruses and other microorganisms. Usually, clinical laboratories isolate total RNA from specimens, although mRNA is actually used as the template by reverse transcriptase.
Kits are available for mRNA extraction only. However, mRNA constitutes only about 1% to 4% of the total RNA of a cell, so it is generally easier to isolate total RNA and use that for RT-PCR. Automated nucleic acid extraction systems can be used to isolate RNA. The integrity of the RNA template can be assessed by agarose gel electrophoresis, UV spectrophotometry, or the use of proper controls during the RT-PCR assay.
Generally, RNA extracted with the use of commercial kits is of high quality, so most clinical laboratories use controls to assess the quality and integrity of RNA specimens. Genomic DNA in a specimen may contaminate the RNA template, so laboratories should treat RNA with a DNase-an enzyme that hydrolyzes DNA—before the RNA is used in RT-PCR. Many RNA extraction kits include a DNase step.
Internal controls are commonly used during RT-PCR. When clinical laboratories assay for RNA viruses from human clinical specimens, an internal control for a human RNA species is generally used to assess the integrity and quality of a specimen.
Internal controls for these assays include transcripts from human genes such as β-actin, glyceraldehyde phosphate dehydrogenase (GAPDH), 18S rRNA, and others. Each clinical specimen must be positive during an RT-PCR assay for one of these internal controls or the assay is not valid.
PCR products that result from RT-PCR assays may be assayed in clinical microbiology laboratories by agarose gel electrophoresis or by real-time PCR. As for standard PCR, most laboratories now use real-time PCR to analyze RT-PCR amplicons.
Some commercially available kits include all the components used for RT-PCR assays, and most of these are designed for real-time PCR or are readily amenable to real-time PCR. In addition, some kits and ASRs are available for the detection of particular organisms by RT-PCR.
Two general types of RT-PCR assays are used by clinical and research laboratories:
One-step RT-PCR: One-step RT-PCR uses a single tube to conduct the reverse transcriptase step and subsequent PCR cycling of the cDNA
Two-step RT-PCR: In Two-step RT-PCR, the reverse transcription of the RNA template is performed first. Once completed, the amplification of the cDNA is carried out in a separate reaction.
Advantages of using one-step RT-PCR
- One of the key advantages of using one-step RT-PCR is that it minimizes the potential carryover of amplicons to the working environment, equipment, and other assay reagents.
- Tube to tube variation is reduced because potential errors are not induced by removing amplicon from the first tube and pipetting into other tubes.
Advantage of two-step RT-PCR
- An advantage of two-step RT-PCR is that resulting cDNA can be used in many different types of subsequent reactions, especially if a laboratory attempts to optimize a reaction.
- Two-step RT-PCR is also useful for the detection of more than one type of transcript from a sample.
- For example, cDNA template could be removed from the first tube and added to a second tube, with primers specific for one type of transcript.
- At the same time, the same cDNA template could be added to a different second tube, with other primers specific for a different transcript.
- Use of two-step RT-PCR requires careful pipetting; clinical laboratories should use a flow hood to reduce
the risk of carryover when opening the first cDNA tube.
Steps of RT-PCR
- The initial step of RT-PCR entails synthesizing a cDNA complementary to RNA transcript with reverse transcriptase. This is not a specific reaction; cDNA is synthesized from all transcripts in a tube.
- Several types of reverse transcriptase are available, including avian myeloblastosis virus (AMV) reverse transcriptase, Moloney murine leukemia virus reverse transcriptase, and rTth reverse transcriptase from Thermus thermophilus, among others.
- Many commercial reverse transcriptases are blended enzymes that have different properties, including enzymes that have reverse transcriptase and DNA polymerase features.
- Thus, the same enzyme can be used in one-tube RT-PCR. Many reverse transcriptases function best at 42° C, so the first step of many RT-PCR assays is incubation at 42° C for 30 minutes.
- Following this step, the temperature is raised to 95° C for 1 to 5 minutes to denature DNA and, for several commercial enzymes, inactivate the reverse transcriptase function of the enzyme.
- Raising the temperature also activates the DNA polymerase function of blended enzymes.
- Standard PCR cycling is then performed and the PCR products are analyzed.
Uses of RT-PCR
- RT-PCR is often used in clinical microbiology laboratories to detect RNA viruses from clinical specimens.
- RT-PCR can be used to quantify the amount of viruses in clinical specimens as well; this is performed for HIV and hepatitis C virus with the Roche Amplicor System.
- Other applications of RT-PCR include quantitative analysis of gene expression, detection of human genes involved in diseases, and detection of cancers from human specimens.
Reference and Sources
- https://cmr.asm.org/content/20/1/49
- https://en.wikipedia.org/wiki/Reverse_transcriptase
- https://en.wikipedia.org/wiki/Reverse_transcription_polymerase_chain_reaction
- https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/reverse-transcription-polymerasechain-reaction
- https://www.promega.in/resources/guides/nucleic-acid-analysis/pcr-amplification/
- https://www.researchgate.net/publication/327283205_Control_Selection_for_RNA_Quantitation
Also Read:
- Types of microscopes
- Different Types of chromatography
- Microscopy: Overview, Principles and Its Types
- Biosafety Cabinet: Introduction, Development and Safety guidance
- Transcription in prokaryotes: Initiation, Elongation and termination
- Chromatography: Introduction, Principle, Classification and Applications
- Microbiology Disciplines: Bacteria, Viruses, Fungi, Archaea and Protists
- Mutations: Introduction, Types, Causes and Repair Mechanisms
- Different types of Pathways for ATP Production
- Electrophoresis: Overview, Principles and Types
- Third Golden Age of Microbiology
