Microbial Identification and Strain Typing Using Molecular Techniques
Introduction
With the rise of large numbers of antibiotic resistance isolates of many microorganisms, increases in the rates of toxin-producing bacteria, and spread of pathogenic microbes worldwide, there is a need for accurate epidemiologic surveillance of these organisms.
Standard techniques in clinical microbiology laboratories do not often provide the necessary resolution to distinguish types and strains from each other. Instead, more refined techniques that separate related organisms from each other at the genetic level are necessary for epidemiologic investigations.
Many typing methods have been described to determine genetic relatedness among bacteria, fungi, and parasites, and some provide better resolution than others. These techniques, often called DNA or genetic fingerprinting, are based on the mutations that accumulate in biologic organisms over time.
Strain typing techniques are often used to compare local strains to determine whether a local outbreak is caused by a single strain type or to multiple strains. A single strain that is responsible for a disease outbreak may have a point source that can then be targeted by the local public health service. Strain typing methods are also used to compare local isolates with worldwide isolates, which can show long-term spread of a strain or strains (clonality).
Whether testing local isolates or isolates from many locales, the technique chosen should have high discriminatory power or the ability to finely resolve different strains. Genetic fingerprinting techniques involve nonamplified and amplified methods.
Non-amplified strain typing
Nonamplified strain typing methods usually involve the analysis of restriction enzyme fragments of chromosomal DNA. As strains diverge genetically over time, restriction enzyme sites on the chromosomal DNA will also change as point mutations accumulate.
When chromosomal DNA from different strains is digested with one restriction enzyme and separated by agarose gel electrophoresis, fragments of different sizes will be observed. These fragments are referred to as restriction fragment length polymorphisms (RFLPs). Comparisons of these RFLP patterns lead to strain typing, and evolutionary ancestors can be derived from the RFLP patterns of strains. Similar RFLP patterns imply genetic relatedness.
Non-amplified strain typing
Nonamplified typing techniques include Southern blotting, plasmid profile analysis, restriction enzyme analysis of chromosomal DNA, pulsed field-gel electrophoresis (PFGE), and multilocus enzyme electrophoresis (MLEE).
Amplified DNA fingerprinting methods use PCR. Known primers or arbitrary primers are used, depending on the particular technique. Commonly used amplified methods include arbitrarily primed PCR (AP-PCR), also called random amplified polymorphic DNA (RAPD), repetitive palindromic extragenic elements PCR (Rep-PCR), and multilocus sequence typing (MLST).
Nonamplified Typing Methods
Southern Blotting
Southern blotting, is sometimes used in epidemiologic investigations to analyze RFLP patterns. Chromosomal DNA is digested with a restriction enzyme, and the resulting fragments are separated by agarose gel electrophoresis. These fragments are transferred from the agarose gel to a nylon or nitrocellulose membrane, and a labeled probe is used to identify a specific target. One of the most popular targets is the DNA that codes for rRNA in microorganisms.
When rRNA RFLP patterns are detected by Southern blotting, the technique is called ribotyping. The genes that code for rRNA are conserved in different species of organisms and appear in conserved positions of a species chromosome. Therefore, ribotyping displays excellent reproducibility and discriminatory power.
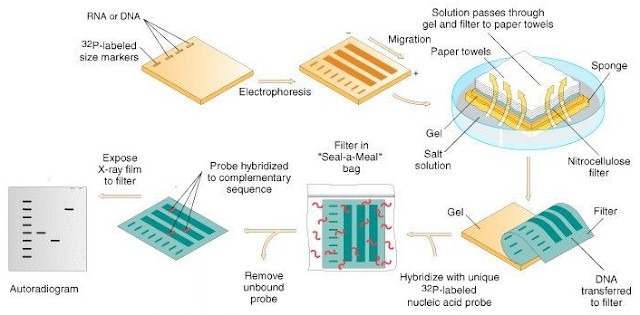
A fully automated ribotyping platform that uses Southern analysis methodology is the RiboPrinter Microbial Characterization System . The instrument automates the entire process, including lysing cells and data processing. Unknown patterns are compared with known patterns to identify strains. Some reports have indicated that other methods, such as PFGE and RAPD, have greater discriminatory power than automated ribotyping.
However, automated ribotyping is advantageous for high-volume laboratories that evaluate large numbers of isolates. Other targets for Southern blot analysis are insertion sequences and transposons, which often contain genes involved in virulence and antibiotic resistance. However, insertion sequences are not present in all strains of bacteria, so some strains are nontypeable.
Plasmid Profile Analysis
This technique was one of the first methods used to type strains of bacteria. Plasmids are extrachromosomal, circular pieces of DNA found in variable number in the cytoplasm of many bacteria. Many bacteria carry antibiotic resistance genes, virulence genes, and other targets of interest on plasmids.
The theory behind this technique is that a given strain with a plasmid profile will be unique from other strains with other plasmids. When a plasmid profile of different isolates is the same, it is possible that these strains are identical.
Plasmid DNA is extracted from bacteria being investigated and separated by agarose gel electrophoresis. Unfortunately, because plasmids are extrachromosomal genetic elements, they are readily lost by bacteria. Some plasmids also contain transposons, or so-called jumping genes, that are readily transferred to other bacteria.
In addition, plasmids can exist in different forms in bacterial cells. Plasmids that have been cut or nicked will have different sizes on agarose gels than uncut plasmids. All these factors can readily change the plasmid profile of a given bacterial isolate, so this technique is not often used and is not considered very reproducible.
Restriction Enzyme Analysis of Chromosomal DNA
This technique is similar to Southern blotting. Chromosomal DNA is extracted from isolates of interest and digested with a restriction enzyme (or more than one restriction enzyme). The enzyme used for digestion should cut the chromosomal DNA in several places so that small and large fragments are produced.

The resulting RFLP pattern is analyzed by agarose gel electrophoresis; transfer to a membrane and subsequent use of a probe to identify a specific sequence is not performed. The discriminatory power of this method is not high, and many of the fragments overlap and are difficult to distinguish from each other.
Pulsed-Field Gel Electrophoresis
Schwartz and Cantor developed PFGE in the early 1980s. The technique is extremely popular and is perhaps the most well-known epidemiologic investigation method. It also may be the most used of the strain typing and identification techniques. A restriction enzyme is used to digest chromosomal DNA at a small number of sites along the chromosome. This results in large DNA fragments that are difficult to resolve by standard agarose gel electrophoresis.

During PFGE, these large fragments are separated in a low-percentage, low-melt agarose gel by an angled electrical field that periodically changes orientation. These changes in electrical field orientation are pulses that successfully force the large DNA fragments through the agarose gel and separate them by size. A low-percentage agarose gel is used for the large DNA fragments, and low-melt agarose enables large fragments to migrate easier.
Relatively simple RFLP patterns result from this process, so comparisons are usually easy. PFGE is also very reproducible and can theoretically be used on any organism. Special equipment that can provide electrical pulses is necessary to run PFGE.

Generally, a system must be obtained specifically for PFGE. Most PFGE assays take several hours (up to overnight) to separate the large DNA fragments produced in the restriction enzyme reaction efficiently. Most PFGE systems incorporate a chilling module within the system to cool the electrophoresis buffer and circulate it to improve separation.
Without circulation and a chilling unit, the buffer will become very warm when separation proceeds for several hours, warm enough potentially to degrade the low-melt agarose gels used in PFGE. The warm temperatures also can produce wavy and difficult to interpret RFLP patterns. Among the various PFGE systems available is the GenePath Strain Typing System (Bio-Rad).
Multilocus Enzyme Electrophoresis
Unlike the other typing techniques described in this section, MLEE analyzes gene expression polymorphism. Analysis of protein polymorphisms by electrophoresis has been studied since the 1970s. MLEE involves the extraction of proteins from isolates of interest, followed by electrophoretic separation and selective staining of these proteins.
The expression of the protein’s genotype is reflected in the position of the stained band, according to the protein’s mobility. Mobility is determined by the net charge of the protein and by the structure of the protein. Two bands of the same protein in different positions after separation suggests two different conformations of the protein, two alleles of the same gene.
MLEE is considered to be an excellent strain typing method. Unfortunately, the DNA sequence of the proteins separated during MLEE cannot be directly assumed because different DNA sequences can result in the same protein because of the redundancy of the genetic code.
In addition, two completely different proteins could have the same mobility and might be interpreted to be the same proteins. Another problem is that interlaboratory comparisons are difficult. Because of these potential problems with MLEE, MLST was eventually developed.
Amplified Typing Methods
Random Amplified Polymorphic DNA
The RAPD technique, also called arbitrarily primed PCR, was first described in 1990 by two different groups, Welsh and McClelland and Williams and colleagues. This is a popular method of DNA fingerprinting. Small primers (≈10 nucleotides in length) with random sequences are used during RAPD.
These primers thus do not have a specific target. Instead, the random primers indiscriminately amplify chromosomal DNA during PCR cycles, which results in fragments of varying lengths after separation by agarose gel electrophoresis. A particular strain will have a different fragmentation pattern than other strains.

RAPD is a simple DNA fingerprinting method capable of providing high resolution and discriminatory power, particularly when more than three different random primers are used. A single primer can be used in a RAPD PCR assay because the single random oligonucleotide can prime random targets on either strand of template DNA.
However, the discriminatory power of just one primer is low with RAPD. The discriminatory power increases if three or more primers are used, although this increases the assay time. Many laboratories use eight or more primers for
RAPD DNA fingerprinting.
One potential problem of RAPD is interlaboratory agreement and reproducibility; this method typically provides excellent intralaboratory reproducibility. RAPD often yields some minor amplicons that exhibit low reproducibility in the same laboratory. Some researchers believe that PFGE has higher discriminatory power than RAPD analysis.
Repetitive Palindromic Extragenic Elements Polymerase Chain Reaction
The Rep-PCR technique is an amplification strain typing method first described in 1991 by Versalovic and associates. All organisms have repetitive DNA sequences—the repetitive palindromic extragenic elements—that repeat throughout the genome.
The unique DNA sequences that lie in between these palindromic repeats are amplified during Rep-PCR using primers specific for the repeat DNA. Rep-PCR results in fragments of varying size, depending on the locations of the palindromic repeats. The amount of DNA in between the repeats varies from strain to strain.
The discriminatory power of Rep-PCR is thought to be a little lower than for PFGE, although results seem to correlate well between the methods; some studies suggest that Rep-PCR has superior discriminatory power. The technique is easy to use and can be scaled up for several isolates, although RAPD assays are somewhat easier to perform.
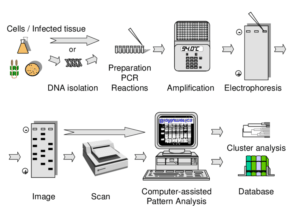
As always, a given laboratory should evaluate the methods to determine the most feasible DNA fingerprinting procedure for the number of expected isolates and for the type of equipment and expertise available.
The DiversiLab System (Bacterial Barcodes, Athens, GA) is an automated rep-PCR instrument on the market that uses proprietary rep-PCR primers for different organisms (e.g., C. difficile and S. aureus). The system uses a microfluidic chip that evaluates rep-PCR products.
As many as 13 different isolates can be evaluated on one chip. Rep-PCR product fragments are analyzed with
the Internet-based DiversiLab software (bioMerieux) to determine strain relatedness. Strain patterns can also be compared with the patterns obtained by other laboratories that use the DiversiLab system.
Multilocus Variable Number of Tandem Repeat Analysis
Like Rep-PCR, multilocus variable number of tandem repeat analysis (MLVA) takes advantage of repetitive DNA sequences in genomes. MLVA amplifies regions of DNA that contain repeats. Repeated sequences in genomes tend to be unstable, so a given bacterial strain may have more repeats at one locus than a different bacterial strain of the same species.
When there are different numbers of repeated sequences at a given locus, it is referred to as a variable number of tandem repeats (VNTRs). MLVA maps VNTRs among bacterial strains by using PCR. This is a useful typing method because it produces quantitative data and is easy to perform, reproducible, and easy to interpret.
Multilocus Sequence Typing
Multilocus sequence typing (MLST) is a derivation of MLEE, described in 1998 by Maiden and coworkers, that analyzes the sequences of genes. Specifically, MLST is used to identify alleles by determining the internal sequences of housekeeping genes. Housekeeping genes are genes that code for proteins necessary for basic functions for cells. Housekeeping genes are constitutive genes (i.e., they are almost always turned on), so they are almost always being expressed.
Both eukaryotic and prokaryotic organisms have housekeeping genes. Human housekeeping genes include the β-actin gene and the glyceraldehyde-3-phosphate dehydrogenase gene. Bacterial housekeeping genes include the 16S rRNA gene and dihydrofolate reductase. Because housekeeping genes are almost always turned on, they also make excellent controls for many molecular methods.
For a typical MLST assay, several loci are chosen that represent different internal regions of housekeeping genes. The PCR assay is used to amplify the DNA at each locus; primers are designed to act as complement to highly conserved regions of the housekeeping genes. Once amplicon is obtained, it is sequenced with an automated sequencer and given a unique number based on the sequence.

Strains of a particular species can then be compared using the same loci. High resolution between strains is possible by comparing these DNA sequences, or the numbers representing these sequences. In addition, the method has achieved excellent interlaboratory comparisons; other laboratories merely need to use published primers for loci of the same species to compare data and determine the spread of strains.
The method has been achieving global recognition, and there is an Internet-based database of strain type, the Multi Locus Sequence Typing home page. The Web site provides protocols, information, and software for sequence analysis. A drawback of MLST is that the laboratory requires an automated sequencer, and the method is expensive compared with PFGE. However, of all the methods described in this chapter for DNA fingerprinting of strains, MLST probably has the highest resolution and the greatest chance of interlaboratory agreements.
Reference and sources
- http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1413-86702003000100005
- https://jcm.asm.org/content/54/3/593
- https://www.researchgate.net/publication/242780567_Interpreting_Chromosomal_DNA_Restriction_Patterns_Produced_by_Pulsed-Field_Gel_Electrophoresis_Criteria_for_Bacterial_Strain_Typing
- https://www.researchgate.net/publication/7984263_Clinical_Evaluation_of_the_DiversiLab_Microbial_Typing_System_Using_Repetitive-Sequence-Based_PCR_for_Characterization_of_Staphylococcus_aureus_Strains
- https://www.sciencedirect.com/topics/neuroscience/pulsed-field-gel-electrophoresis
- https://www.researchgate.net/publication/272399376_Pseudomonas_aeruginosa_-_Pathogenesis_and_Pathogenic_Mechanisms
- https://www.sciencedirect.com/science/article/pii/019567019190053B
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC87445/
- https://sfamjournals.onlinelibrary.wiley.com/doi/pdf/10.1111/j.1365-2672.2007.03632.x
- https://www.ncbi.nlm.nih.gov/pmc/articles/pmid/23857503/
Also Read:
- what is microbiology?
- What is Gene Expression?
- DNA replication in prokaryotes
- Histones types and its functions
- Proteomics: Introduction, Methods, Types and Application
- Vector: properties, types and characteristics
- Different types of Pathways for ATP Production
- Measurements of microbial growth
- Reverse Transcription Polymerase Chain Reaction (RT-PCR)
- Whole-Genome Shotgun Sequencing: overview, steps and achivements
- DNA Replication in eukaryotes: Initiation, Elongation and Termination
- Mutations: Introduction, Types, Causes and Repair Mechanisms
- CRISPR-Cas9 Gene editing tool: Introduction, Principles, Uses & Applications
- Whole-Genome Shotgun Sequencing: overview, steps and achievements
