The Genetic Code – Overview
The genetic code is the set of rules by which the sequence of nucleotides in DNA or RNA is translated into the sequence of amino acids in proteins. It is universal, triplet-based, and highly conserved across almost all living organisms.
The final step in the expression of protein-coding genes is translation. Protein synthesis is called translation because it is a decoding process. The information encoded in the language of nucleic acids must be rewritten in the language of proteins. During translation, the sequence of nucleotides is “read” in discrete sets of three nucleotides, each set being a codon.
Each codon codes for a single amino acid. The sequence of codons is “read” in only one way-the reading frame to give rise to the amino acid sequence of a polypeptide. Deciphering the genetic code was one of the great achievements of the twentieth century. Here we examine the nature of the genetic code.
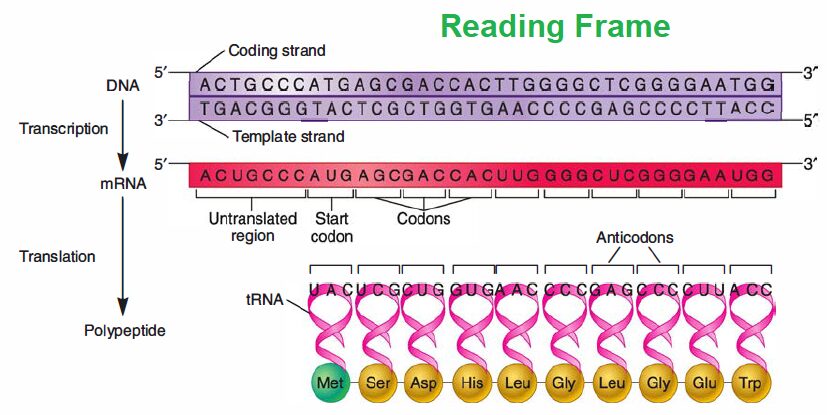
The genetic code, presented in RNA form. Close inspection of the code reveals several features that are related not only to the way cells use DNA to store information but also to why it is valuable for storing data, as described in the chapter opening story. One feature is that the code words (codons) are three letters (bases) long; thus one small “word” conveys a significant amount of information. Each codon is recognized by an anticodon present on a tRNA molecule.

Another feature is that the code has “punctuation:’ One codon, AUG, is almost always the first codon in the protein-coding portion of mRNA molecules. It is called the start codon because it serves as the start site for translation by coding for the initiator tRNA. Three other codons (UGA, UAG, and UAA) are involved in termination of translation and are called stop or nonsense codons.
These codons don’t encode an amino acid and thus don’t have a tRNA bearing their anticodon. Thus only 61 of the 64 codons in the code, the sense codons, direct amino acid incorporation into protein. Finally, the genetic code exhibits code degeneracy; that is, there are up to six different codons for a given amino acid.
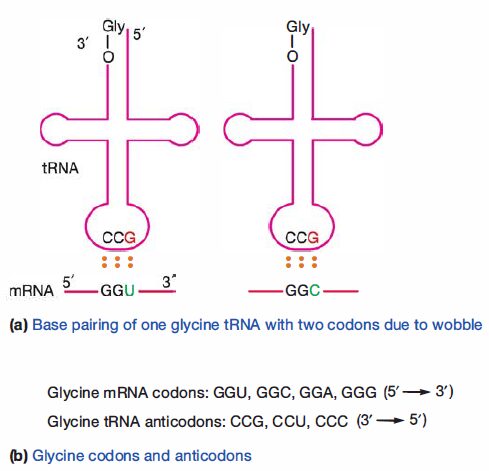
Despite the existence of 61 sense codons, there are fewer than 61 different tRNAs. It follows that not all codons have a corresponding tRNA. Cells can successfully translate mRNA using fewer tRNAs because loose pairing between the 5′ base in the anticodon and the 3′ base of the codon is tolerated. Thus as long as the first and second bases in the codon correctly base pair with an anticodon, the tRNA bearing the correct amino acid will bind to the mRNA during translation. This is evident on inspection of the code.
Note that the codons for a particular amino acid most often differ at the third position. This somewhat loose base pairing is known as wobble, and it relieves cells of the need to synthesize so many tRNAs. Wobble also decreases the effects of some mutations.
The description of the genetic code just provided is of the universal genetic code. However, there are exceptions to the code. The first exceptions discovered were stop co dons that encoded one of the 20 amino acids. For instance, some protists have a single stop codon (UGA); the other two stop codons are recognized by tRNAs bearing glutamine (Gln).
Some Exceptions to the Universal Genetic Code
| Codon | Amino Acid Inserted | Where Observed |
| AGA and AGG | Stop | Mammalian mitochondria |
| AGA and AGG | Serine (Ser) | Invertebrate mitochondria |
| AUA | Methionine (Met) | Mammalian, invertebrate, and yeast mitochondria |
| CUA | Threonine (Thr) | Yeast mitochondria |
| CUG | Serine | Some fungi |
| UAA and UAG | Glutamine (Gln) | Some protists |
| UAG | Pyrrolysine | Some methanogens and bacteria |
| UGA | Selenocysteine | Members of all three domains |
| UGA | Tryptophan (Trp) | Mammalian, invertebrate, and yeast mitochondria |
| UGA | Glutamine | Mycoplasma bacteria |
More dramatic deviations from the code have also been discovered. Proteins from members of all three domains of life have been discovered to contain the amino acid selenocysteine, the twenty-first amino acid. Pyrrolysine, the twenty-second amino acid, can be found in the proteins of several methanogens and at least one bacterium.

Genomic analysis indicates that pyrrolysine might also exist in many other bacteria and some eukaryotes. Selenocysteine is inserted at certain UGA codons, whereas pyrrolysine is inserted at UAG codons.
Reference and Sources
- https://en.wikibooks.org/wiki/Cognitive_Psychology_and_Cognitive_Neuroscience/Print_version
- https://www.lecturio.com/magazine/dna-transcription-and-translation/
- https://quizlet.com/279412091/microbiology-test-3-material-flash-cards/
- https://quizlet.com/165922180/chapter-13-transcription-translation-flash-cards/
- https://quizlet.com/206191016/final-micro-205-flash-cards/
Also Read:
- Nitrogen Cycle
- Transposable Elements
- Vector: properties, types and characteristics
- DNA Replication in eukaryotes: Initiation, Elongation and Termination
- Transcription in prokaryotes: Initiation, Elongation and Termination
- Proteomics: Introduction, Methods, Types and Application
- Plasmid: Properties, Types, Replication and Organization
- Milk: Composition, Processing, Pasteurization, Pathogens and Spoilage
- Cider: Production, Extraction, Fermentation and Maturation
- Fungi: Distribution, Morphology, Reproduction, Classification
- CRISPR-Cas9 Gene editing tool: Introduction, Principles, Uses & Applications


8 thoughts on “The Genetic Code”
Good information avariable in this page keep post like this information it is very useful to us
Thank you Swetha keep supporting..