Introduction
Proteomics is the large-scale study of proteins, particularly their structure, function, expression, and interactions within a biological system. It complements genomics and is crucial for understanding cellular processes, disease mechanisms, and drug discovery. Proteomics is the systematic analysis of the entire set of proteins (proteome) expressed by a cell, tissue, or organism under specific conditions.
- Genome function can be studied at the translation level as well as the transcription level. The entire collection of proteins that an organism produces is called its proteome. Thus proteomics is the study of the proteome or the array of proteins an organism can produce.
- Proteomics provides information about genome function that mRNA studies cannot because a direct correlation between mRNA and the pool of cellular proteins does not always exist. Much of the research in this area is referred to as functional proteomics.
- It is focused on determining the function of different cellular proteins, how they interact with one another, and the ways in which they are regulated.
Methods used in proteome analysis
- Two-dimensional gel electrophoresis
- SDS polyacrylamide gel electrophoresis (SDS-PAGE)
Two-dimensional gel electrophoresis
- In this procedure, a mixture of proteins is separated using two different electrophoretic procedures (dimensions). This permits the visualization of thousands of cellular proteins that would not otherwise be separated in a single electrophoretic dimension. The first dimension makes use of isoelectric focusing, in which proteins move through a pH gradient (e.g., pH 3 to 10).
- First, the protein mixture is applied to an acrylamide gel in a tube with a stable pH gradient and electrophoresed. Each protein moves along the pH gradient until the protein’s net charge is zero and the protein stops moving. The pH at this point is equal to the protein’s isoelectric point. Thus the first dimension separates proteins based on the content of ionizable amino acids.

SDS polyacrylamide gel electrophoresis (SDS-PAGE)
- The second dimension is SDS polyacrylamide gel electrophoresis (SDS-PAGE). SDS (sodium dodecyl sulfate) is an anionic detergent that denatures proteins and coats them with a negative charge.
- After the isoelectric gel has been completed, the tube gel is soaked in SDS buffer and then placed at the edge of an SDS-PAGE gel. A voltage is then applied. Because all the proteins are negatively charged thanks to SDS treatment, they will migrate from the negative pole of the gel to the positive. In this way, polypeptides are separated according to their molecular weight i.e., the smallest polypeptide will travel fastest.
Application of Proteomics
- Two-dimensional gel electrophoresis can resolve thousands of proteins; each protein is visualized as a spot of varying intensity, depending on its cellular abundance.
- Radiolabeled proteins are often used, enabling greater sensitivity so that newly synthesized proteins can be distinguished.
- Computer analysis is used to compare two-dimensional gels from microbes grown under different conditions or to compare wild-type and mutant strains. Websites have been developed for the deposition of such images, allowing researchers access to ever-growing databases.
Importance of mass spectrometry in analyzing protein structure
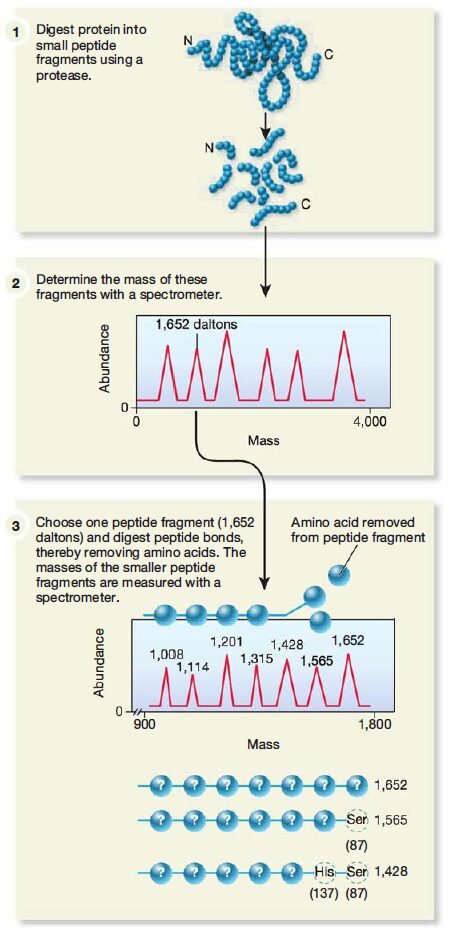
- Two-dimensional gel electrophoresis is even more powerful when coupled with mass spectrometry (MS). The unknown protein spot is cut from the gel and the protein is cleaved into fragments by treatment with proteolytic enzymes. Then the fragments are analyzed by a mass spectrometer and the mass of the fragments is plotted. This mass fingerprint can be used to estimate the probable amino acid composition of each fragment and tentatively identify the protein.
- Sometimes proteins or collections of fragments are run through two mass spectrometers in sequence, a process known as tandem MS. The first spectrometer separates proteins and fragments, which are further fragmented. The second spectrometer then determines the amino acid sequence of each smaller fragment.
- The sequence of a whole protein often can be determined by analysis of such fragment sequence data. Alternatively, if the genome of the organism has been sequenced, only a partial amino acid sequence is needed.Computer analysis is then used to compare this amino acid sequence with the predicted translated sequences of all the annotated CDSs on the organism’s genome. In this way, both the protein and the gene that encodes it can be identified.
Types of Proteomics
- A second branch of proteomics is called structural proteomics. Here the focus is on determining the three-dimensional structures of many proteins and using these to predict the structures of other proteins and protein complexes. The assumption is that proteins fold into a limited number of shapes and can be grouped into families of similar structures.
- When a number of protein structures are determined for a given family, the patterns of protein structure organization or protein-folding rules will be known. Then computational biologists use this information to predict the most likely shape of a newly discovered protein; a process known as protein modeling.
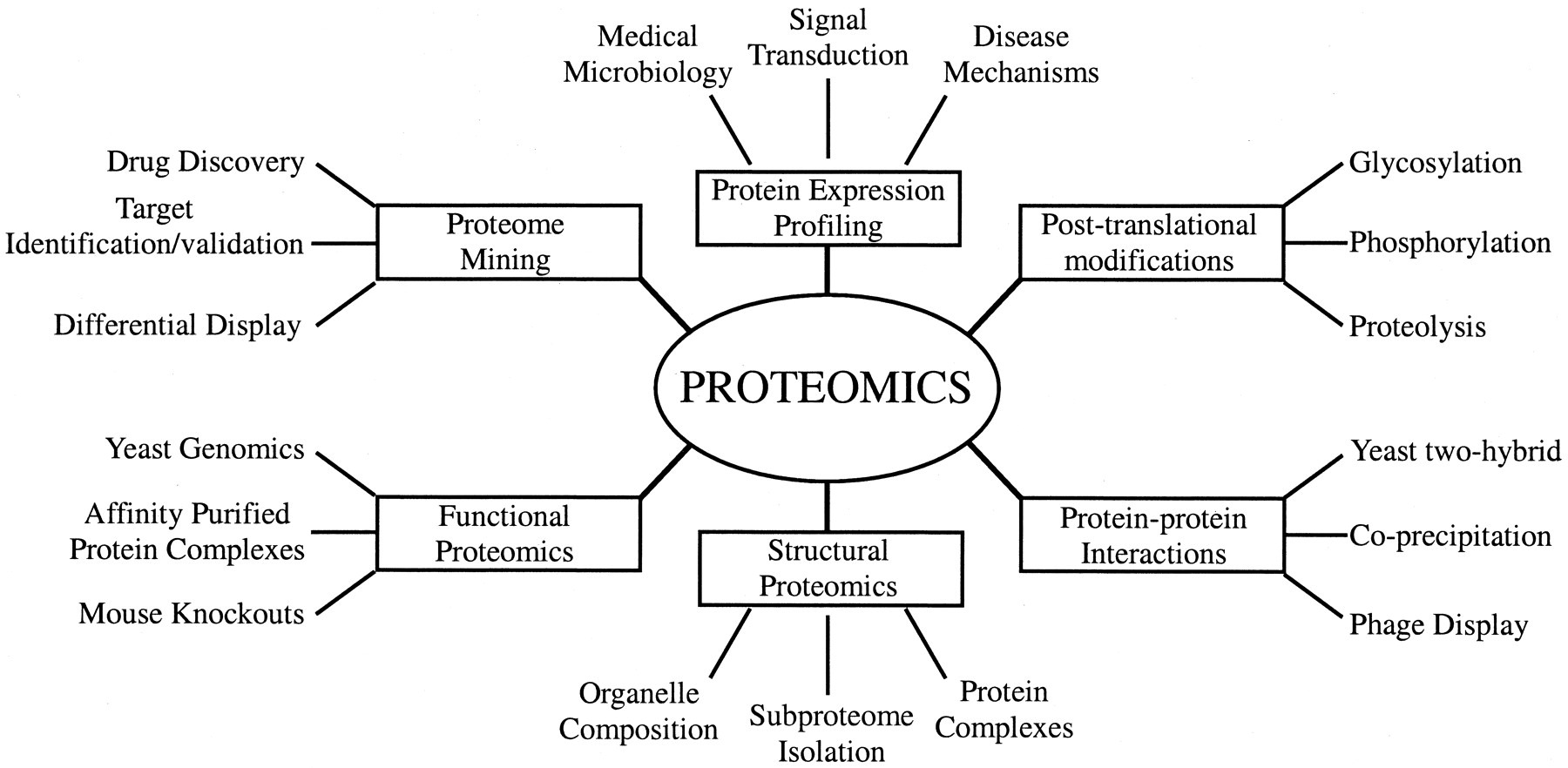
- A variety of other “-omics” are studied. For example, lipidomics investigates a cell’s lipid profile at a particular time, and glycomics is the systematic study of the cellular pool of carbohydrates.
- The goal of metabolomics is to identify all the small-molecule metabolites, regardless of chemical structure, present in the cell at a given point in time. Because many metabolites are used in multiple pathways, the overview presented by a complete metabolome enables a researcher to discern which pathways are functional under a given set of conditions.
- Thus metabolomics provides a high-resolution means to evaluate physiological status. These “-omics” rely heavily on the use of chromatography and as such are beyond the scope of this chapter.
- Nonetheless, the development of these fields illustrates the dynamic and ever-changing nature of microbiology and the need to take a holistic view if one seeks to understand the structure, function, and behavior of cells.
Reference and Sources
- https://quizlet.com/71004695/chapter-18-micro-flash-cards/
- https://pubs.niaaa.nih.gov/publications/arh26-3/219-232.htm
- https://quizlet.com/249421143/final-exam-flash-cards/
- https://en.wikipedia.org/wiki/Proteomics
- https://dokumen.pub/molecular-and-cell-biology-sos-sie-0070153795-9780070153790.html
- https://www.deepdyve.com/lp/wiley/from-protein-to-the-beginnings-of-clinical-proteomics-bCZMdrNTRk
- https://en.wikipedia.org/wiki/Expressed_sequence_tag
- https://courses.lumenlearning.com/boundless-biology/chapter/genomics-and-proteomics/
- https://quizlet.com/81234364/ls3-midterm-1-flash-cards/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6267353/
Also Read:
- Whole-Genome Shotgun Sequencing: overview, steps and achievements
- Milk: Composition, Processing, Pasteurization, Pathogens and Spoilage
- Polymerase Chain Reaction
- Southern Blotting
- Vector: properties, types and characteristics
- Granulocytes: Introduction, Types, Functions and Roles
- Measurements of microbial growth

