Influenza (Flu)
Introduction
Influenza (Italian, un influenza di freddo-to be influenced by the cold), or the flu, was described by Hippocrates in 412 BCE. The first well-documented global epidemic of influenza-like disease occurred in 1580. Since then, 31 possible influenza pandemics have been documented, with four occurring in the twentieth century. The worst pandemic on record occurred in 1918 and killed approximately 50 million people around the world.
This disaster, traced to the Spanish influenza virus, was followed by pandemics of Asian flu (1957), Hong Kong flu (1968), Russian flu (1977), and the novel swine flu (2009). (The names reflect popular impressions of where or from what the episodes began, although all are now thought to have originated in China.) Another new influenza strain erupted from pigs in 2011, an H3N2. Health officials are keeping an eye on it.
Organelles of the secretory and endocytic pathways
Influenza is a respiratory system disease caused by negative strand RNA viruses that belong to the family Orthomyxoviridae. There are three genera that cause significant human disease:
- Influenza virus A
- Influenza virus B
- Influenza virus C
Influenza viruses are widely distributed in nature, infecting a variety of mammal and bird hosts.

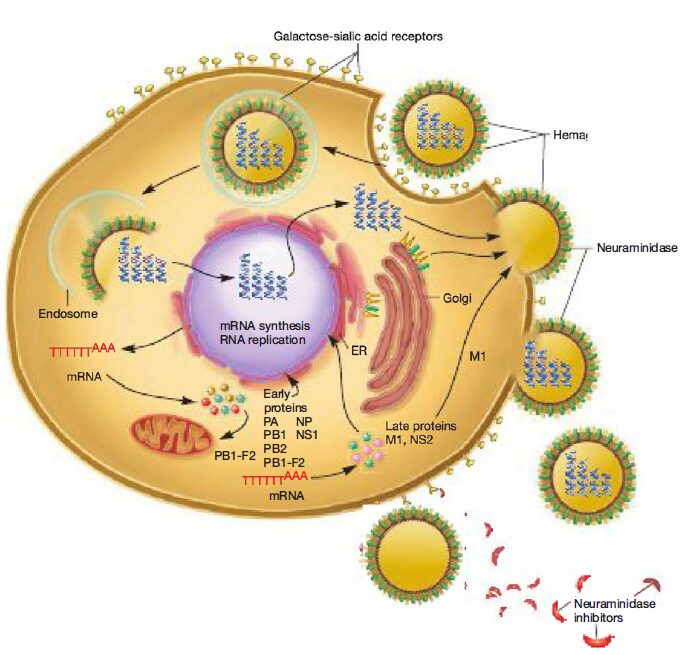
All influenza viruses are acquired by inhalation or ingestion of virus contaminated respiratory secretions (thus the public health recommendation to “cover your cough and wash your hands frequently”). During an incubation period of 1 to 2 days, viral particles adhere to cells of the host respiratory epithelium by their hemagglutinin spike proteins.
Neuraminidase present in other envelope spikes of influenza types A and B may assist in virus adherence by hydrolyzing the mucus that covers the epithelium. Influenza virus attachment triggers receptor mediated endocytosis, and encloses the virion in an endosome.
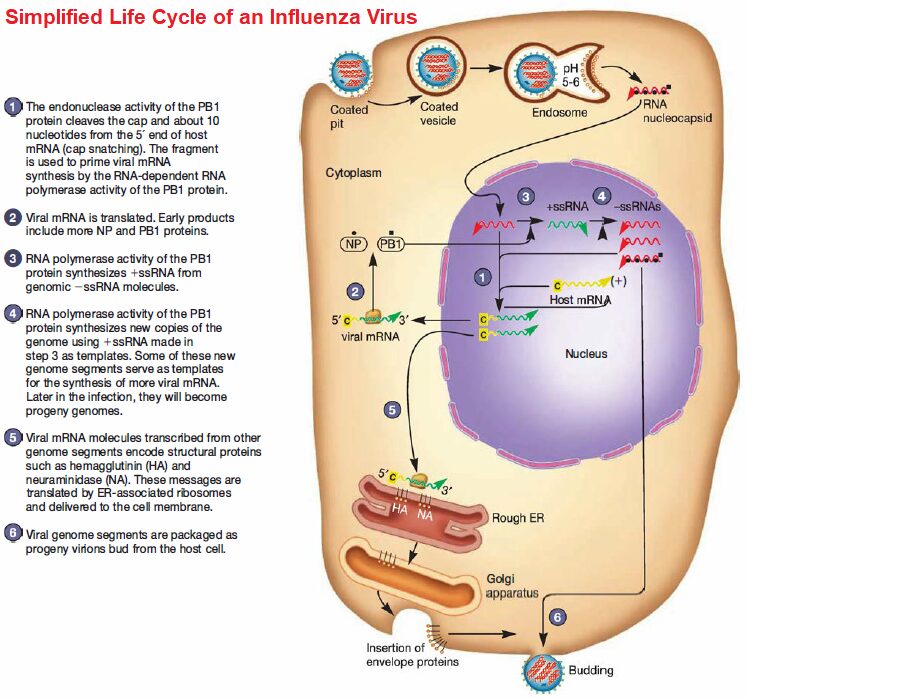
The hemagglutinin molecule in the virus’s envelope undergoes a dramatic conformational change when the endosomal pH decreases. The hydrophobic ends of the hemagglutinin spring outward and extend toward the endosomal membrane. After they contact the membrane, fusion occurs and the RNA nucleocapsid is released into the cytoplasm.
Patterns of infectious disease in a population
Influenza viruses contain seven to eight segments (pieces) of linear RNA, with a total genome length between 12,000 to 15,000 nucleotides. Each RNA segment encodes one or two proteins. Replication and transcription of viral RNAs is accomplished in the host cell nucleus by three polymerase subunits and the nucleoprotein.
Newly synthesized viral ribonucleo proteins are then exported from the nucleus to the cytoplasm, where they assemble into virions at the host cell plasma membrane. The mature virions are released from the host cell, taking host membrane as the viral envelope. The enveloped virion can be spherical (80 to 120 nm in diameter) or filamentous (200 to 300 nm long, 20 nm in diameter).

Types of Influenza viruses
Influenza viruses are classified into subtypes (or strains) based on their membrane surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). There are 16 HA (H1-H16) and 9 NA (N1-N9) antigenic forms known; they recombine to produce the various HA/NA subtypes. As mentioned previously, HA and NA function in viral attachment and virulence.
Specifically, the HA protein is required for binding of individual virions to host cell receptors and the subsequent fusion of the viral envelope with the host endosomal membrane, initiating infection. The NA protein facilitates release of newly assembled influenza virions from infected cells by cleaving sialic acid residues between virus and host membranes. Thus drugs that inhibit NA prevent newly formed virions from being released from the host cell membrane; they stay attached and cannot infect other host cells.
Together HA and NA protein efficiency defines the virulence of influenza viruses by facilitating virus attachment to susceptible host cells and mediating virus shedding, respectively. Host specificity can be explained in part by the difference in receptor binding. Human influenza viruses preferentially attach by way of their HA protein to receptors on human tracheal epithelial cells. In contrast, avian influenza viruses preferentially bind receptors found on intestinal epithelial cells of waterfowl (the main viral replication site of avian influenza viruses), for the most part, restricting infection to birds.
However, the genomes of the influenza viruses are significantly plastic in that they change frequently due to point mutations and reassortment of the RNA segments. This is noteworthy, because it leads to the sudden appearance of new influenza viruses that cause pandemics.
Genome plasticity is one of the most important features of influenza viruses in that it leads to both subtle and sometimes drastic changes in the viral proteins that are synthesized, altering virus antigenicity. If the protein variation is small, it is called antigenic drift. Antigenic drift results from the accumulation of mutations of HA and NA in a single strain of flu virus within a geographic region.

This usually occurs every 2 to 3 years, causing local increases in the number of flu infections. Antigenic shift is a large antigenic change resulting from the reassortment of genomes when two different strains of flu viruses (from both animals and humans) infect the same host cell and are incorporated into a single new capsid.
Because there is a greater change with antigenic shift than antigenic drift, antigenic shifts can yield major epidemics and pandemics. Antigenic variation occurs almost yearly with influenza A virus, less frequently with the B virus, and has not been demonstrated in the C virus, thus the need for yearly influenza immunization.
Animal reservoirs are critical to the epidemiology of human influenza. For example, rural China is one region of the world in which chickens, pigs, and humans live in close, crowded conditions. Influenza is widespread in birds; although birds cannot usually transmit the virus to humans, they can transfer it to pigs. Pigs can transfer it to humans and humans back to pigs. Recombination between human and avian strains thus occurs in pigs, leading to novel HA/NA combinations causing major antigenic shifts. To track the various influenza subtypes, a standard nomenclature system was developed.
It includes the following information: group (A, B, or C), host of origin, geographic location, strain number, and year of original isolation. Antigenic descriptions of the HA and NA surface proteins are given in parentheses for type A viruses only. The host of origin is indicated for viruses obtained from nonhuman hosts; for example, A/swine/Iowa/15/30 (H1N1). However, the host origin is not given for human isolates; for example, A/Hong Kong/03/68 (H3N2).
Influenza A viruses having H1, H2, and H3 HA antigens, along with N1 and N2 NA antigens, are predominant in nature, infecting humans since the early 1900s. H1N1 viruses appeared in 1918 and were replaced in 1957 by H2N2 viruses as the predominant subtype. The H2N2 viruses were subsequently replaced by H3N2 as the principle subtype in 1968.
The H1N1 subtype reappeared in 1977 and cocirculates today with H2N1, H3N2, H5N2, H7N2, H7N3, H7N7, H9N2, H10N7, and H5N1 viruses. Importantly, the receptor-binding specificity of human and avian influenza viruses just discussed suggests that in order to infect humans, surface HA and NA proteins of avian influenza viruses must change so as to be able to recognize human-type receptors.
Indeed, genome sequences of the earliest isolates of the 1918, 1957, and 1968 pandemics demonstrated that the otherwise avian influenza A viruses possessed HAs that recognized specific receptors with a characteristic structure (SA-α2,6-Gal) usually seen in human influenza.
Of significance is the fact that, while the world was expecting the H5N1 subtype (also known as bird flu) of influenza A to be the next cause of an influenza pandemic, a novel H1N1 subtype of influenza A evolved and quickly spread around the world. Epidemiological data evaluating large numbers of influenza and pneumonia cases in La Gloria, Veracruz, Mexico, and California during mid-February 2009 (the end of the seasonal flu season) suggested that a new strain of flu was the cause.
The novel H1N1 (previously referred to as swine flu) outbreak was quickly confirmed by Mexican and U.S. public health officials. On April 26, 2009, the United States declared a public health emergency, implementing the nation’s pandemic response plan. The World Health Organization (WHO) also sent out several alerts that called for heightened levels of response as the international spread of the disease continued. Forty-one countries reported 11,034 cases, including 85 deaths by May 21, 2009. On June 11, 2009, the WHO raised the pandemic alert level again, indicating that a global pandemic was in progress with more than 70 countries reporting cases of H1N1 infection with ongoing community-level outbreaks in multiple parts of the world.
The WHO reported 99,103 cases ofH1N1 with 476 fatalities on July 6, 2009. Ten days later, officials at the WHO indicated that the counting of individual cases was no longer essential for monitoring either the level or nature of the risk posed by the pandemic virus or to guide implementation of the most appropriate response measures. As a result, routine counting and reporting of novel H1N1 cases was halted except for newly affected areas.
By June 19, 2009, all 50 states, the District of Columbia, Puerto Rico, and the U.S. Virgin Islands reported novel H1N1 cases. The commencement of the regular influenza season in the Southern Hemisphere complicated the detection and bspread of novel H1N1, making accurate counts ofH1N1 infection impossible.
So although the CDC reported 43,771 confirmed cases and 302 deaths by July 24, 2009, officials believed that more than a million Americans had been sickened by the virus. By this time, the majority ofH1N1 cases were still occurring in younger people, with the median age reported to be twelve to seventeen years, based on data from Canada, Chile, Japan, the United Kingdom, and the United States.
Some reports suggest that patients requiring hospitalization and those with fatal illness may be slightly older and have underlying disease, such as asthma. Cardiovascular disease, respiratory disease, diabetes, and cancer were also considered risk factors for serious H1N1 disease with fatal complications. H1N1 vaccines became available in October 2009 with the goal to use them in addition to the 2009 seasonal flu vaccine. Limited supply of the H1N1 vaccine by the end of November 2009 prevented this.
The CDC estimated that between April and October 2009, there were as many as 34 million H1N1 infections, with an approximate 153,000 hospitalizations and upward of 6,100 deaths. The epidemic was declared over on May 22, 2010. The death toll was approximately 10,000, far less than the worst-case projections.
The novel H1N1 (swine) flu pandemic of 2009 diverted attention from the continuing threat of the highly pathogenic H5N1 (avian) flu. The H5N1 subtype was responsible for six deaths in 1997, although it had been sporadically detected prior to that. Throughout 2003 to 2008, however, H5N1 was responsible for a substantial number of bird infections, resulting in the culling of millions of birds, even though no significant human-to-human transmission had occurred.
The birds were destroyed to help prevent transmission of the virus to susceptible humans. Yet by the end of 2011, H5N1 was responsible for 516 confirmed cases of influenza and 341 deaths in 9 countries. The case fatality
rate differs between countries and genders (women have a higher death rate) but exceeds 60%. Importantly, despite the high mortality
rate, humans are rarely infected.
Because of potentially severe consequences of pandemic H5N1 infection of humans, an international effort coordinated by the WHO continues to monitor virus surveillance, epidemiology, and control efforts for this virus subtype, as well as for H1N1. A single-dose H5N1 influenza A vaccine has been developed, using inactivated, whole-virion antigens, by researchers in Hungary and the United Kingdom. It stimulates protective immunity in volunteers and fulfills the strict criteria for licensing in the United States. However, it is ineffective against the most recent H 5N1 influenzaA strain that is circulating through out Asia.
Antiviral drugs
Zanamivir and oseltamivir are chemically related antiviral drugs known as neuraminidase inhibitors; they have activity against both influenza A and B viruses. Neuraminidase inhibitors prevent the release of newly formed virions from their host cell by blocking the catalytic site of the enzyme neuraminidase. Newly formed virions exit their host cell by budding.
However, the final step of release occurs when neuraminidase cleaves the sialic acid residue on a host membrane receptor that is attached to the virus’s hemagglutinin spike. With the neuraminidase blocked, virions are not detached from the host cell membrane and therefore cannot travel to infect another cell. Importantly, aspirin (salicylic acid) should be avoided in children younger than fourteen years to reduce the risk of Reye’s syndrome.
Vaccines and immunization
The main stay for prevention of influenza since the late 1940s has been inactivated virus vaccines, especially for the chronically ill, individuals over age sixty-five, residents of nursing homes, and health-care workers in close contact with people at risk. Clinical disease in these patients is most likely to be severe. Because of influenza’s high genetic variability, efforts are made each year to incorporate new virus
subtypes into the vaccine. Even when no new subtypes are identified in a given year, annual immunization is still recommended because immunity using the inactivated virus vaccine typically lasts only 1 to 2 years.
Reference and Sources
- https://accessmedicine.mhmedical.com/content.aspx?sectionid=94110540
- https://www.chegg.com/homework-help/questions-and-answers/animal-critical-epidemiology-influenza-foundmany-parts-rural-china-poultry-pigs-humans-li-q58709587
- https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0008495
- https://www.health.com/condition/infectious-diseases/worst-pandemics-in-history
- https://www.cdc.gov/h1n1flu/updates/061209.htm#:~:text=On June 11, 2009, the World Health
Organization,cases of human infection with novel H1N1 flu. - https://www.cambridge.org/core/journals/epidemiology-and-infection/article/an-epidemiological-study-ofinfluenza-viruses-among-chinese-farm-families-with-household-ducks-andpigs/BDAEE9E00FAF005AC92D4046A3458177
- https://www.ncbi.nlm.nih.gov/genome/viruses/variation/help/flu-help-center/influenza-virus-biology/
- https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccinestandardization/influenza
- https://www.chegg.com/homework-help/questions-and-answers/genomes-influenza-virus-change-frequentlydue-mutations-rna-segments-sudden-appearance-new-q58709627
- https://www.cdc.gov/flu/weekly/weeklyarchives2008-2009/weekly28.htm
- https://pubmed.ncbi.nlm.nih.gov/21106732/
- https://quizlet.com/344793245/microbio-chpt-38-flash-cards/
- https://www.reuters.com/article/idUSLG309223
- https://en.wikipedia.org/wiki/Antigenic_drift
