DNA replication in prokaryotes
DNA replication is a very important and complex process in living organisms upon which all life depends. At the time of DNA replication, the double helix strand of DNA are separated and each of them involves for the synthesis of a complementary strand according to the base-pairing rules.
Each of the two progeny DNA molecules contains one new strand and one old strand, and this type of DNA replication is termed as semi-conservative.
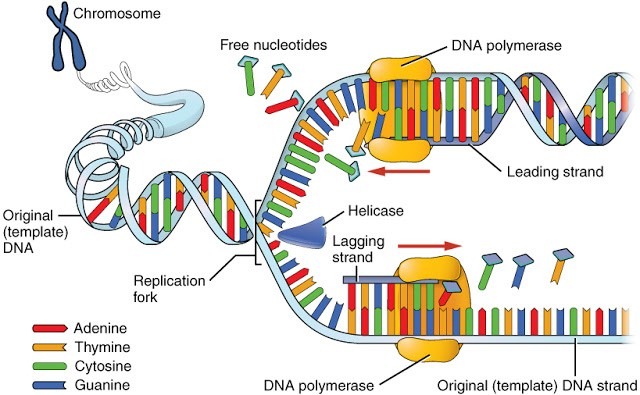
DNA Replication Machinery
DNA replication is essential to organisms, and a great deal of effort has been devoted to understanding its mechanism. The replication of E. coli DNA requires at least 30 proteins. The process of DNA replication is similar in most of the organisms.
Enzymes called DNA polymerases catalyze DNA synthesis. All known DNA polymerases catalyze the synthesis of DNA in the 5′ to 3′ direction, and the nucleotide to be added is a deoxynucleoside triphosphate (dNTP ).
Deoxynucleotides are linked by phosphodiester bonds formed by a reaction between the hydroxyl group at the 3′ end of the growing DNA strand and the phosphate closest to the 5′ carbon (the a-phosphate) of the incoming deoxynucleotide.

The energy needed to form the phosphodiester bond is generated by the release of the terminal two phosphates as pyrophosphate (PPi) from the nucleotide that is added.
The PPi is subsequently hydrolyzed to two separate phosphates (Thus the deoxynucleoside triphosphates dATP, dTTP, dCTP, and dGTP serve as DNA polymerase substrates while deoxynucleoside monophosphates (dNMPs: dAMP, dTMP, dCMP, dGMP ) are incorporated into the growing chain.
For DNA polymerase to catalyze the synthesis of DNA, it needs three things.
- The first is a template, that is used to direct the synthesis of a complementary DNA strand.
- The second is a primer to give a free 3′-hydroxyl group to which nucleotides can be added.
- The third is a set of dNTPs. E. coli has five different DNA polymerases (DNA polymerase I-V). DNA polymerase III plays the important role in replication.
DNA polymerase III holoenzyme
DNA polymerase III holoenzyme is a multifunctional enzyme composed of 10 different proteins. Most evidence suggests that within the complex are found two core enzymes Each core enzyme binds one strand of DNA and is responsible for catalyzing DNA synthesis and proof reading the product to ensure fidelity of replication.
The 13 clamp tethers a core enzyme to the DNA. At the middle of the holoenzyme, is look like an octopus like structure which has a complex of proteins known as the clamp loader, which is important for loading the 13 clamp onto DNA.

A dimer of another protein (tau) holds the holoenzyme together. Because there are two core enzymes, both strands of DNA are bound by a single DNA polymerase III holoenzyme. DNA polymerase III holoenzyme is merely one component of a huge complex of proteins called the replisome.
Other proteins found in the replisome include helicases, single-stranded DNA binding proteins, and topoisomerases.
Helicases are liable for separating (unwinding) the DNA strands just before the replication fork, using energy from ATP hydrolysis.
Single-stranded DNA binding proteins (SSBs) keep the strands apart once they separated.
Topoisomerases relieve the strain or tension generated by the rapid unwinding of the double helix (the replication fork may rotate as rapidly as 75 to 100 revolutions per second).
Once the template is prepared, the primer needed by DNA polymerase III can be synthesized. An enzyme called primase synthesizes short RNA strands, mainly around 10 nucleotides long and complementary to the DNA, This serves as the primer.
Steps in the Replication Fork
In E. coli, DNA replication is initiated at specific nucleotides called the oriC locus (for origin of chromosomal replication). This site is AT rich. Recall that adenines pair with thymines using only two hydrogen bonds, so AT-rich segments of DNA become single stranded more readily than do GC-rich regions. This is important for initiation of replication.
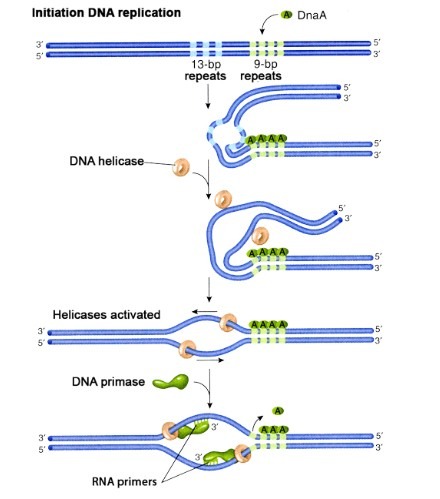
- The initiator protein for Bacteria DnaA is mainly important for initiating DNA replication. DnaA proteins bind regions in oriC throughout the cell cycle, but to initiate replication, DnaA proteins must bind a few particular oriC sequences. The presence of DnaA at these sites recruits a helicase (usually DnaB helicase) to the origin.
- The helicase unwinds the helix with the aid of topoisomerases such as DNA gyrase. The single strands are kept separate by SSBs.
- Primase synthesizes RNA primers as needed. A single DNA polymerase III holoenzyme catalyzes both leading strand and lagging strand synthesis from the RNA primers.
- After most of the lagging strand has been synthesized by the forming of Okazaki fragments, DNA polymerase 1 seperate the RNA primers.
- DNA polymerase I does this because, unlike other DNA polymerases, it has the ability to snip off nucleotides one at a time starting at the 5′ end while moving toward the 3′ end of the RNA primer.
- Finally, the Okazaki fragments are joined by the enzyme DNA ligase, which forms a phosphodiester bond between the 3 ‘-OH of the growing strand and the 5 ‘-phosphate of an Okazaki fragment.

Termination of Replication
In E. coli, DNA replication can stops when the replisome reached to a termination point on the DNA.
A protein known as Tus binds to the ter sites and halts the formation of the forks. In many different types of bacteria, replication halt spontaneously when the forks meet.
Regardless of how fork movement is stopped, there is mainly two problems that should be solved by the replisome. One is the forming of interlocked chromosomes called catenanes.
The other is a dimerized chromosome in which two chromosomes joined each other to form a single chromosome twice as long.
Catenanes are formed when topoisomerases breaks and rejoin the DNA strands to release the supercoiling of the replication fork.
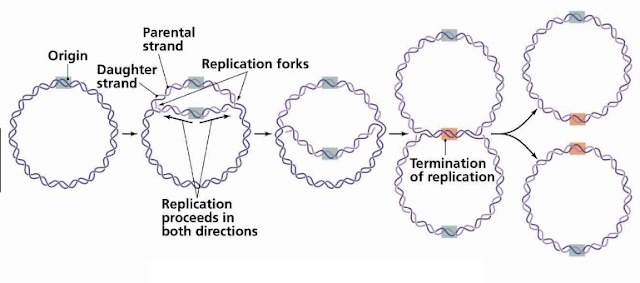
The two daughter DNA molecules are separated by topoisomerases that break both strands of one molecule, pass the other DNA molecule synthesis occurs at each replication fork.
An enzyme called telomere resolvase (ResT) cuts the two chromosomes apart as it forms hairpin ends for each daughter molecule.
Reference and Sources
- 1% – https://alex-aufpassen.com/content/139/3/45e-k13150-xrvn1
- 1% – https://homepages.rpi.edu/~bellos/dna_replicn’.htm
- 1% – https://www.sciencedirect.com/topics/nursing-and-health-professions/deoxyribonucleoside-triphosphate
- 1% – https://tophat.com/marketplace/science-&-math/biology/textbooks/oer-openstax-biology-openstax-content/79/4106/
- 1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC136941/
- 2% – https://quizlet.com/328769463/chapter-13-bacterial-genome-replication-expression-flash-cards/
- 1% – https://wikimili.com/en/DNA_polymerase_III_holoenzyme
- 1% – https://schoolbag.info/test/mcat_1/5.html
- 1% – https://www.researchgate.net/publication/6845405_Spatial _and_temporal_organization_ of_the_Bacillus_subtilis_replication_cycle
- 1% – https://www.studyblue.com/notes/note/n/micro-3813-study-guide-2013-14-callaghan/deck/8690770
- 1% – https://opentextbc.ca/microbiologyopenstax/chapter/dna-replication/
- 1% – https://www.sciencedirect.com/topics/neuroscience/dna-polymerase-iii-holoenzyme
- 1% – https://openoregon.pressbooks.pub/mhccmajorsbio/chapter/dna-replication-in-prokaryotes/
- 1% – https://pubs.acs.org/doi/10.1021/bi500019s <1% – https://www.golifescience.com/enzymes-involved-in-dna-replication/
- 1% – https://www.slideshare.net/rajasekhar342/dna-replication-in-prokaroytes-and-in-eukaryotes
- 1% – https://course-notes.org/book/export/html/243
- 1% – https://en.wikipedia.org/wiki/Replisome
Also Read:
- What is Gene Expression?
- DNA Replication in eukaryotes: Initiation, Elongation and Termination
- Transcription in prokaryotes: Initiation, Elongation and Termination
- Polymerase Chain Reaction: Introduction, working and Components
- Plasmid: Properties, Types, Replication and Organization

I need notes for different topics –of genetics and molecular biology–mutations,dna damage and repair .
Ok very soon will start uploading notes on these topic
Thanks for comments..
Good
thankss bro keep supporting
I need notes about insulin, hGH and Factor VIII