DNA Replication in eukaryotes
Content
- Introduction
- DNA Replication in Eukaryotes
- Initiation of DNA Replication in Eukaryotes
- Eukaryotic Primases and DNA Polymerases
- Telomeres and Telomerases
Introduction
- DNA replication is similar in all cellular organisms. It is accomplished by a huge complex of proteins called the replisome.
- Central to the functioning of the replisome are the DNA polymerases responsible for leading and lagging strand replication.
- All DNA polymerases catalyze the synthesis of DNA in the 5′ to 3′ direction using a template to synthesize a complementary DNA strand and a primer to provide a free 3′-hydroxyl group to which nucleotides can be added.
- The DNA of most bacteria is circular and replication begins at a single point, the origin of replication.
- The bacterial replicative DNA polymerase (DNA polymerase III) is recruited to the origin only after the initiator protein DnaA begins assembly of the bacterial replisome, which is composed of at least 30 proteins.
- The replication forks move bidirectionally from the origin until the entire circular chromosome is replicated.
DNA Replication in Eukaryotes

- The distinctive features of eukaryotic DNA replication arise from differences in chromosome structure and the replication machinery.
- Eukaryotes have multiple chromosomes, each of which is usually much larger than a typical bacterial chromosome.
- The DNA in eukaryotic chromosomes is linear, which means that a mechanism for replicating chromosome ends is needed.
- Finally, eukaryotic DNA is wound around histones. These must associate with the newly synthesized strand of DNA.
Initiation of DNA Replication
- Replication of eukaryotes is initiated at multiple origins of replication. This allows the chromosome to be replicated much faster than it could be if there were only one origin per chromosome.
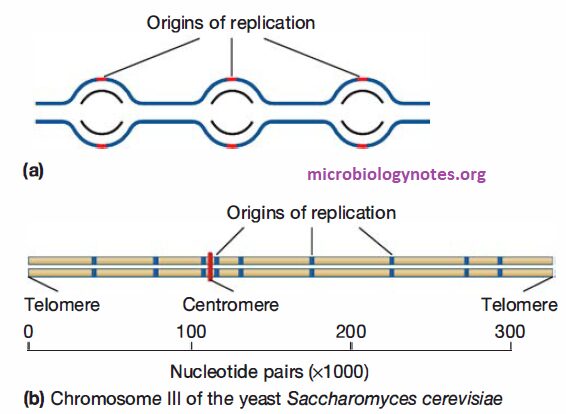
- Two replication forks move outward from each origin until they encounter replication forks that formed at adjacent origins.
- Thus eukaryotic chromosomes consist of multiple replicons, rather than the single replicon (i.e., the entire chromosome) observed in bacteria.
- In eukaryotes, origins of replication are “marked” by a complex of proteins called the origin recognition complex (ORC), which remains bound to the origins throughout much of the cell cycle.
Major Replisome Proteins in Bacteria and Eukaryotes
| Function | Bacteria | Eukaryotes |
| Initiator; binds origin | Dna A | ORC |
| Helicase loader | DnaC | Cdc6 and Cdt1 |
| Helicase | DnaB | MCM complex |
| Single-strand binding | SSB proteins | RPA |
| Primer synthesis | DnaG primase | Pol α-primase1 |
| Replicative DNA polymerase | DNA polymerase III (C-family polymerase) | DNA polymerase δ and DNA polymerase ε (B-family polymerases) |
| Clamp loader | ϒ complex | RF-C |
| Clamp | β clamp | PCNA |
| Ligase | DNA ligase2 | DNA ligase 12 |
| Primer removal | Ribonuclease H; DNA polymerase I | RNaseH/Fen1 |
- Six different ORC proteins have been identified (Orc1 to Orc6), and these are combined in a species-specific manner to form the ORC.
- For instance, the ORC of the yeast Schizosaccharomyces pombe consists of six ORC proteins, two each of Orc1 (origin-recognition-complex protein 1), Orc2, and Orc4.
- ORC serves as a platform on which other proteins assemble in a cell cycle-dependent fashion. The first two proteins to associate with ORC are Cdc6 (cell-division-cycle protein 6) and Cdtl (CdclO-dependent-transcript 1 protein).
- The two proteins bind ORC in the late M/early G 1 phases of the cell cycle.
- Together, ORC, Cdc6, and Cdtl recruit a set of proteins called the MCM complex to the origin, thereby forming the pre-replicative complex (pre-RC). Pre-RCs are activated as the cell cycle transitions from the Gl phase to the S phase by phosphorylation of Cdc6.
- Cdc6 and Cdtl are then replaced by numerous other proteins, forming the complete replication machinery. At this point, MCM, which has helicase activity, begins unwinding DNA and replication begins.
Eukaryotic Primases and DNA Polymerases
- DNA polymerases from many organisms have been analyzed and sorted into seven families based on their amino acid sequence and structure.
DNA Polymerase Families
| Family | Example(s) | Functions/Comments |
| A | Bacterial DNA polymerase I | Replacement of RNA primers present in Okazaki fragments |
| B | Archaeal DNA polymerase B (Pol B) Eukaryotic DNA polymerases α, δ, and ε |
Replicative DNA polymerases |
| C | Bacterial DNA polymerase III | Replicative DNA polymerase |
| D | Archaeal DNA polymerase D (Pol D) | Replicative DNA polymerase in some archaea; unique to archaea |
| X | Eukaryotic DNA polymerase β | DNA repair |
| Y | Bacterial DNA polymerase IV (Din B) | DNA repair |
| Reverse transcriptase (RT) | Retroviral reverse transcriptases Telomerase RT | RNA-dependent DNA polymerase |
- As shown in table eukaryotic DNA polymerases are found in different families than are the bacterial enzymes.
- In eukaryotes, three DNA polymerases are responsible for DNA replication:
- DNA polymerases α-primase
- DNA polymerases ε-primase
- DNA polymerases δ-primase
- Synthesis of the leading and lagging strands begins with the formation of a primer in all organisms, regardless of domain.
- However, among eukaryotes, this is accomplished with a bifunctional enzyme called DNA polymerase α-primase (often called simply Pol α-primase).
- This enzyme consists of four subunits:
- Two enzyme catalyze DNA synthesis (Pol α).
- Two enzyme catalyze RNA synthesis (primase).
- The primer is made in two steps:
- The primase component of the enzyme makes a short RNA strand (∼10 nucleotides), which is then transferred to the active site of Pol α.
- Pol α adds an additional 20 or so deoxyribonucleotides to the RNA strand. Thus in eukaryotes, the single-stranded primer for DNA replication is an RNA-DNA hybrid molecule.
- Once the primer is formed, the other two DNA polymerases take over.
- Evidence suggests that DNA polymerase ε (Pol ε) is responsible for leading-strand synthesis, whereas DNA polymerase δ (Pol δ) carries out lagging-strand synthesis.
- Thus unlike most bacteria where DNA polymerase III synthesizes both leading and lagging strands, two distinct DNA polymerases carry out these functions in eukaryotes.
Telomeres and Telomerases: Protecting the Ends of Linear DNA Molecules
- The fact that the DNA in eukaryotic chromosomes is linear poses several problems.
- Without protection, the ends are susceptible to degradation by enzymes called DNases.
- They are also able to fuse with the ends of other DNA molecules, thus generating aberrant chromosomes, which may not segregate properly during cell division.
- Finally, linear chromosomes present a problem during replication because of DNA polymerase’s need for a primer that provides a free 3′-0H.
- When the primer for the Okazaki fragment at the end of the daughter strand is removed, the daughter molecule is shorter than the parent molecule.
- Over numerous rounds of DNA replication and cell division, this leads to a progressively shortened chromosome.
- Ultimately the chromosome loses critical genetic information, which could be lethal to the cell. This problem is called the “end replication problem,” and a cell must solve it if it is to survive.

- Eukaryotic cells have solved the difficulties related to having linear chromosomes by forming complex structures called telomeres at the ends of their chromosomes and by using an enzyme called telomerase.
- Telomeres are protein-DNA complexes that protect the linear DNA within them from degradation and end fusion.
- The protein component of a telomere varies from species to species, as does the length of DNA present.
- Telomeric DNA contains many copies of a particular sequence of nucleotides, placed one after the other (tandem repeats).
- Importantly, the DNA is single-stranded at the very end and is called the G-tail because it is rich in guanosine bases; telomerase uses the G-tail to maintain chromosome length.
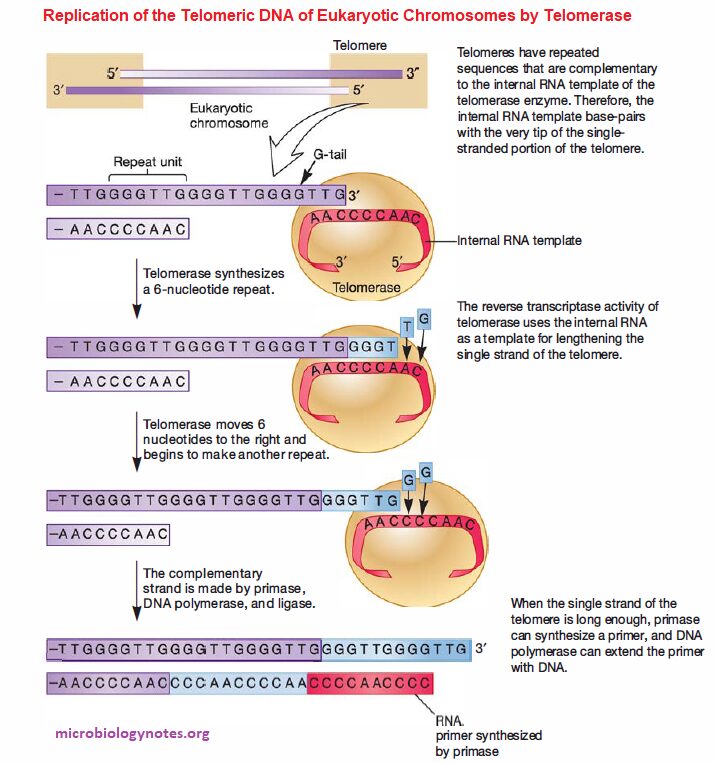
Telomerase has two important components:
-
- An internal RNA template
- An enzyme called telomerase reverse transcriptase (RT).
An internal RNA template
- The internal RNA template is complementary to a portion of the G-tail and base-pairs with it.
- The internal RNA template provides the template for DNA synthesis, which is catalyzed by telomerase RT (i.e., the 3′ -OH of the G-tail serves as the primer for DNA synthesis).
- After being lengthened sufficiently, there is room for synthesis of an RNA primer, and the single strand of telomere DNA can serve as the template for synthesis of the complementary strand. Thus the length of the chromosome is maintained.
Telomerase reverse transcriptase (RT)
- Telomerase RT deserves additional comment. As described in the preceding paragraph, telomerase RT synthesizes DNA using an RNA template.
- Enzymes with this capability are defined as RNA-dependent DNA polymerases.
- RNA-dependent DNA polymerases are not unique to telomerases; certain viruses use RNA-dependent DNA polymerases to complete their life cycles (e.g., human immunodeficiency viruses and hepatitis B viruses).
Reference and Sources
- https://en.wikibooks.org/wiki/An_Introduction_to_Molecular_Biology/Replication_of_DNA_and_its_repair
- https://www.researchgate.net/publication/318570421_Structural_biology_of_telomerase_and_its_interaction_at_telomeres
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3201878/
- https://en.citizendium.org/wiki/Replication_of_a_circular_bacterial_chromosome
- https://www.researchgate.net/publication/5412778_Division_of_Labor_at_the_Eukaryotic_Replication_Fork
- https://www.intechopen.com/books/the-mechanisms-of-dna-replication/telomeres-their-structure-andmaintenance
- https://www.chegg.com/homework-help/questions-and-answers/bacterial-replicative-dna-polymerase-recruitedsingle-origin-initiator-protein-dnaa-begins-q6906805
- https://quizlet.com/416058535/biol-202-week-5-flash-cards/
- https://quizlet.com/279912407/chapter-11-chromosome-structure-and-organelle-dna-flash-cards/
- https://www.sciencedirect.com/science/article/pii/S0022283613006050
Also Read:
- Transcription in prokaryotes: Initiation, Elongation and termination
- Plasmid: Properties, Types, Replication and Organization
- Spectroscopy: Introduction, Principles, Types and Applications
- Vector: properties, types and characteristics
- Bacteriophage: characteristics and replication of lytic and lysogenic cycle
- Chickenpox (Varicella) and Shingles (Herpes Zoster)
- Nitrogen Cycle
