Tryptophan Operon: History, Structure, Regulation and Attenuation
Introduction
The discovery and explanation of the Operon model can be regarded as the milestone in the field of molecular biology. In 1961 Jacob and Monod published an article “Genetic Regulatory Mechanisms in the Synthesis of Proteins” which we now regard as the operon model.
The studies were carried out on the metabolism of lactose in E coli. Later, in 1965 Francois Jacob, Andre Lwoff, and Jacques Monod were awarded with Nobel prize in Physiology and Medicine for their discoveries concerning genetic control of enzymes and virus synthesis.
Definition and structure of operon
Operon can be described as the genes clustered on one chromosome that are transcribed together under the control of the single promoter. The following figure describes the general structure of an operon.

- Promoter: The promoter is located at the upstream of the structural genes and is the site recognized by the RNA Polymerase and initiates transcription.
- Operator: The segment of DNA located in between the structural genes and the promoter. The site that is recognized by the repressor and hence acts as the physical barrier for the RNA polymerase thus controlling the transcription.
- Structural genes: Genes that are involved in making of the protein.
Types of operons
- Inducible: Example Lac Operon.
- Repressible: Example tryptophan operon.
- There is significant difference in inducible and repressible systems.
- In inducible system the inducer interacts with the products of the regulatory i.e., the repressor making it incapable to bind to the operator and thus inefficient to block the transcription.
- While In repressible systems the elements in the cell (corepressor) interacts with the product of the regulatory genes making it efficient to bind to the operator and thus to block the transcription.
Tryptophan operon
- Tryptophan operon was the first repressible operon discovered.
- Tryptophan operon is the cluster of genes that code for the elements essential for the synthesis of tryptophan which is an essential amino acid.
- Here note the difference between Lac operon and the Tryptophan operon abbreviated as the trp operon; Lac operon works for the catabolism of the lactose while the trp operon works for the synthesis of enzymes needed for the synthesis of tryptophan.
- Therefore, trp operon is an anabolic type of operon.
Structure of the tryptophan operon
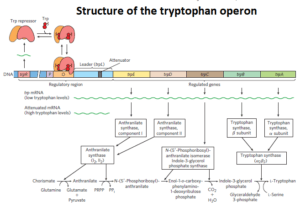
The above figure elaborates the structure of the trp operon from E. coli. The tryptophan operon consists of promoter, operator which overlaps with the promoter followed by the leader sequence of about 162 nucleotides and five structural genes which code for enzymes that are essential for the synthesis of tryptophan.
The five structural genes and their products are as follows:
- trpE : Anthranilate synthase Component I.
- trpD : Anthranilate synthase Component II.
- trpC : N-(5_-Phosphoribosyl)-anthranilate isomeraseIndole-3-glycerolphosphate synthase.
- trpB : Tryptophan synthase β subunit.
- trpA : Tryptophan synthase α subunit.
- The mRNA from the trp operon has the half life period of about 3 min making it efficient to respond to the changing needs of the cell towards this amino acid.
Regulation of tryptophan operon

The regulation of the trp operon takes place through two mechanism viz. the switch ON and switch OFF mechanism and through transcription attenuation.
The switch on and switch off mechanism
- Trpytophan being one of the essential amino acids is being continuously synthesized and hence the usual status of the trp operon is switched ON. Only when the synthesis of the tryptophan is in excess the operon needs to be switched OFF.
- The trp repressor is regarded as the switch of the operon. It is a homodimer. When the tryptophan concentration is exceeds the requirement of the cell it gets associated with the trp repressor causing a conformational change in it. This conformational change in the repressor allows it to bind to the operator thus ceasing the expression of the trp operon.
- As the operator site overlaps with the promoter the binding of the RNA polymerase is also affected by this mechanism.
The transcriptional attenuation mechanism
- Transcriptional attenuation is regarded as the secondary regulatory or fine-tuning regulatory mechanism of the trp operon.
- Transcriptional phenomenon in which the initiation of the transcription takes place normally but the process gets abruptly stopped before the structural genes are transcribed.
- This mechanism utilizes the fact that in prokaryotes the processes of transcription and translation take place simultaneously.
- The attenuation mechanism makes use of the signal encoded in the four sequences within the leader region.
- The regions of the sequence 3 and 4 act as the attenuator. These sequences 3 and 4 are complementary and can form G-C rich stem loop structure followed by series of U residues. This stem loop structure between the sequences 3 and 4 acts s the transcription terminator.
- Sequences 2 and 3 are also are also complementary to each other but the loop structure formed by the 2 and 3 is not capable of ceasing the transcription.
- Regulatory sequence 1 determines the tryptophan sensitive mechanism which decides whether there will be pairing between 2-3 0r 3-4. During translation the regulatory sequence 1 encodes a leader peptide which consists of two Trp residues.
- When the tryptophan concentrations are high the translation proceeds rapidly through the two codons of the tryptophan of the sequence 1 and into the sequence 2 due the abundant availability of the tryptophan.
- In this situation sequence 2 is covered with the ribosome and hence unavailable to base pair with sequence 3 while the sequence 4 is available for pairing thus forming the transcription terminating stem loop structure.
- But when the tryptophan concentration is low the translation of the sequence 1 takes place slowly and the ribosome stops at the two trp residues due to less availability of the tryptophan. Due to this the sequence 2 and sequence 3 are available for pairing with each other.
- But pairing between 2-3 does not affect the transcription as it allows the RNA Polymerase to travel ahead. Thus the tryptophan synthesis remains unaffected and the cell’s tryptophan demand is met.
Reference and Sources
- Lehninger’s Principles of Biochemistry (Edition 6)
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2776167/
- https://www.faunafondness.com/protein-synthesis-regulation/
- https://www.cam.ac.uk/research/research-at-cambridge/nobel-prize
- https://www.sciencedirect.com/science/article/pii/S187456609780010X
- https://europepmc.org/articles/PMC154410/
- https://quizlet.com/453464068/chapter-11-bio-flash-cards/

Where about lac operon?
It is also there.
I wasn’t able to get it. Will you please send me the link or suggest me to find it?